Rice is one of the important food security crops of India. It feeds more than 2.7 billion people across the globe. In spite of best management practices the crop is being affected by a dreaded disease known as Rice blast. This disease is caused by a fungal pathogen Maganporthe oryzae which affects almost all parts of the plant but very damaging during seedling and panicle filling stage (Fig.1). In India, the disease appears where ever rice is grown but more prevalent in the areas where high humidity and low temperature is present during nights. It causes more than 75 % annual yield losses during epidemic conditions. This disease is being managed very effectively by using resistant crop varieties. However, the development of resistant varieties is dependent on the basic knowledge of variability present in the blast fungus population.

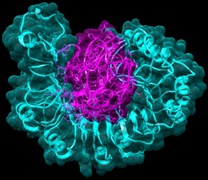
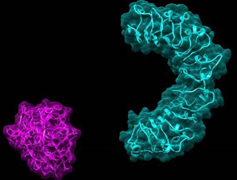
ICAR- NRCPB is actively working on rice-Magnaporthe system for the past more than 16 years and has already cloned and characterized rice blast resistance genes Pi54, Pi54rh and Pi54of from varieties and wild species of rice, besides participated in decoding of rice genome sequence along with International rice genome sequencing project (IRGSP, Nature 2005). However, not much is known about the genome structure of the important rice blast pathogen prevalent in India.
This size of M. oryzae genome is ~38 million base pairs present in six chromosomes. An important strain of M. oryzae present in India was used for decoding its genome sequence with an objective to identify fungal avirulence genes which plays important role in imparting resistance to the host during host-pathogen interaction. A team of scientists lead by Dr. T. R. Sharma, Principal Investigator of the project and Director of NRCPB used Next Generation Sequencing (NGS) method for decoding of complete sequence of an avirulent isolate RML-29 of M. oryzae and predicted 1144 protein coding genes in this pathogen. The results have already been published in an International journal Frontiers in Plant Sciences on August, 2016 and is freely available at http://journal.frontiersin.org/article/10.3389/fpls.2016.01140/full.
Dr Sharma and team reported that they have used a new computational and molecular biology methods to identify the AvrPi54 gene from fungus which interacts with the host gene Pi54 thus impart resistance to the rice plant (Fig.2). This approach has been used for the first time for the identification and cloning of avirulence gene from any plant pathogen. The new method adopted in the study has a great potential to reduce the time and labor involved in identifying and mapping an Avr gene and could be used as an alternative to the more tedious positional cloning approach. The candidate AvrPi54 gene identified and cloned in this study would be helpful in better understanding of the host–pathogen interaction. As the Pi54 gene is already cloned and functionally validated at NRCPB, the newly identified AvrPi54 gene, along with Pi54 gene, could be used in developing broad spectrum resistance against rice blast pathogen.








Like on Facebook
Subscribe on Youtube
Follow on X X
Like on instagram